|
Page 1, 2
Introduction
The last decade and a half has seen the global proliferation of nutrigenetic testing companies. Overall, the direct-to-consumer (DTC) model has been mostly unsuccessful and the growth has come primarily from companies' selling tests through practitioners. Many of the more reputable companies insist on their practitioners' completing an accreditation course before they are "certified" to sell the companies' tests. Although an admirable insistence, the integrity and depth of these courses vary. In addition, practitioners vary in their qualifications and experience. The result is that most practitioners are inadequately trained to interpret and effectively communicate nutrigenetic results and dietary recommendations.
Parallel to this growth in nutrigenetic tests, there has been a welcome increase in nutritional genomics academic programs, ranging from university short courses and elective modules in masters' programs, to postgraduate programs, masters', and doctoral degrees. A number of universities have begun to include a small amount of nutritional genomics teaching in their undergraduate curriculum. Understandably, these programs tend to be research based, and while they potentially offer academic integrity and research skills, they are not helpful in providing practitioners with the skills that they need to work with nutrigenetics in practice.
This article does not address whether or not the field of nutritional genomics is ready for practitioner use. (For this, we recommend "Nutrigenetics and Personalized Nutrition: Are We Ready for DNA-Based Dietary Advice?")1 Rather, it addresses the practitioner "knowledge gap," describing what knowledge practitioners may be missing.
Defining the Knowledge Gap
The Nutrition Knowledge Gap
One of the greatest challenges is in bringing together the fields of nutrition and genetics. For doctors, medical specialists, dentists, homeopaths, and others, there is an acknowledged lack of nutrition education. For those with sufficient training in nutrition such as dietitians and nutritionists, the gap is the understanding of nutrition in a systems biology, functional, and political ecology context.
Nutritionism has become the ruling ideology of our current health care.2 There is a great need for a new nutrition ideology, a new way of seeing and knowing about food, nutrition, health, and conditions such as obesity. Food needs to be taught, spoken about, and practiced not only in a biomedical model but also with due consideration of the social, environmental, political, and economic relations that govern how food is produced and consumed. This requires an understanding of nutrition that starts with genes and ends with the environment; no easy task.
The Nutritional Genomics Knowledge Gap
Many practitioners, and, surprisingly, academics as well, cannot differentiate between nutrigenetics and nutrigenomics (Figure 1). What is often referred to as nutrigenomics is in fact nutrigenetics. In this article we use the term nutritional genomics to mean both nutrigenetics and nutrigenomics. The tests that practitioners work with are more accurately nutrigenetics tests, and the training that they receive usually covers only nutrigenetic interactions (see Defining the Difference Between Nutrigenetics and Nutrigenomics below). This has resulted in an emphasis on SNPs (single nucleotide polymorphisms) and a "superficial" involvement with what the science of nutritional genomics has to offer. It is our contention that practitioners require an understanding of what we call the "three pillars of nutritional genomics" (Figure 2).
These include:
1. nutritional biochemistry
2. nutrigenetics
3. nutrigenomics
Figure 1
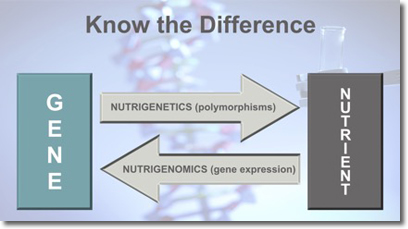
Figure 2

Biochemical processes are driven and understood through nutritional biochemistry. It is imperative to understand the biochemical pathways and the disruption thereof, which contribute to perturbations in health and potentially the development of disease. Nutrigenetics provides information on genetic sequence variants: SNPs as well as other variations such as copy number variants. These may (but not always) alter gene expression or enzyme function. By understanding nutrigenomics (nutrient impact on gene expression), a practitioner is able to identify and advise on nutrients and lifestyle changes that may alter gene expression. Put together, these three pillars of nutritional genomics will empower the practitioner to construct targeted and clinically useful dietary and lifestyle recommendations.
Defining the Difference Between Nutrigenetics and Nutrigenomics
Genetic variability (nucleotide sequence change) influences how we interact with our environment. Nutrigenetics describes the influence of gene variants on our ability to interact with bioactive molecules in the molecular environment surrounding our cells and the consequences of that interaction.
In contrast to nutrigenetic interactions wherein the gene variants act on the environment, with nutrigenomic interactions, the environment influences gene expression. Nutrigenomics is concerned with the influence of bioactive molecules interacting with a gene, potentially influencing gene expression. Either upregulating or downregulating (turning it up or down) or activating or silencing (on or off). Nutrigenomic interactions may be direct or indirect. In a direct interaction, the molecules are of small molecular weight, carrier mediated, and lipid soluble. In an indirect interaction, the molecules are larger and hydrophilic. These molecules interact at the cell surface.
Employing the Three-Pillar Approach in Practice
The Fundamental Strategy
A key consideration in the three-pillar approach is to focus initially on upstream biochemical events. Each gene codes for an enzyme, and that enzyme is involved in one or more biochemical pathways. If practitioners understand the nature of that pathway when it is functioning normally, they can better predict what may happen when a variant of the gene produces an enzyme operating at lower activity. Understanding the function of the key upstream biochemical pathways is at the core of this approach. Human cells utilize a number of core biochemical processes in maintaining their homeostasis; these include redox regulation, detoxification, inflammation modulation, energetics, and methylation. These five upstream processes form the foundation for the first pillar.
Redox dysregulation and associated inflammation as upstream factors are common elements of numerous chronic diseases.3 Nutrigenetic test profiles typically include SNPs that affect the primary antioxidant enzymes and major inflammatory cytokines, whereas they are unlikely to include many of the disease-specific downstream gene variants.
The following example uses type 2 diabetes to examine the evolving identification of associated genes and their function. Genome-wide association studies (GWAS), linkage, and candidate gene association studies have identified a number of diabetes-related genes; however, these studies are inconsistent in their findings. A 2007 study identified the genes PPARG, KCNJ11, and TCF7L2 as established genes associated with common forms of type 2 diabetes.4 Eighteen months later, another study found that the number of gene loci robustly associated with type 2 diabetes had risen from 3 to 18 in the ensuing period.5 Their findings included the obesity-related genes FTO and MC4R, and they concluded that 7 of these identified type 2 diabetes gene loci affect beta-cell function, but the function of 9 other genes was unknown. By 2010, largely through GWAS, approximately 20 gene variants associated with type 2 diabetes had been identified, with some overlap of 10 with body mass index (BMI) and obesity, 4 with fasting glucose levels in the normoglycemic population, and over 30 with lipid levels.6
Clearly, this field is still evolving, and what is clinically significant is that there are no known interventions to modify the expression of some of these genes. In the three-pillar approach, these downstream genes may be considered only after the core upstreambiochemical processes have been addressed.
As with many chronic diseases, redox dysregulation is accepted as a fundamental contributor to both the onset and the progression of type 2 diabetes. Examining the relevant gene variants from a nutrigenetic profile gives the practitioner a guide to the redox-regulating potential of that patient. Following such an analysis, the appropriate nutrigenomic interventions can be selected in order to modify the expression of one or more genes.
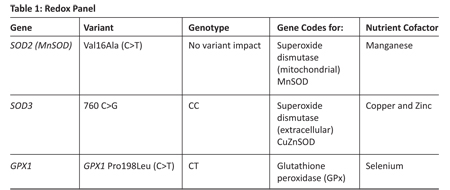
Case Study: A 46-year-old female presents with recently diagnosed type 2 diabetes. Her diet is poor and her BMI is 29. Nutrigenetic testing shows the following SNPs in her redox panel (Table 1). Space does not permit a full nutrigenetic analysis.
Table 1
Pillar 1: The Nutrigenetic Profile
Pillar 2: The Biochemical Analysis
The superoxide radical is generated continuously by the mitochondria from inhaled oxygen. In certain circumstances such as during exercise, there is an increased flux of oxygen through the mitochondria. This increases the quantity of superoxide radical produced and this acts as the prooxidant signal needed to activate the cells' endogenous defense mechanisms. As a consequence, the expression of the MnSOD gene is upregulated so that more MnSOD enzyme is produced; the same applies to increased expression of the GPX gene. Just as exercise increases the flux of oxygen, glucose, and fatty acids through the mitochondria, so too does overeating. Several pathways in type 2 diabetes contribute further to this increased flux .7
These two primary upstreamreactions constitute the first line of cellular defense against oxidative stress.8 As shown in Figure 3, two superoxide radicals dismute to produce hydrogen peroxide, a nonradical reactive oxygen species (ROS) that can readily react with metal ions to produce highly toxic hydroxyl radicals (OH−). If acted upon by GPx in the normal manner, hydrogen peroxide readily converts to unreactive water, thereby removing the oxidative risk.
Figure 3: Superoxide Redox Reaction
Reaction #1: 2O2− + 2h+ SOD H2O2 + O2
Reaction #2: 2 H2O2 GPx 2H2O
In our case study, the patient carries a "normal" MnSOD and so the mitochondria are likely to perform Reaction # 1 adequately, generating hydrogen peroxide. However, she carries a GPx SNP, thereby limiting the activity of the GPx enzyme. As a result, hydrogen peroxide will tend to accumulate at a rate faster than can be handled by the dysfunctional GPx enzyme. Consequently, hydrogen peroxide is more likely to react with available heavy metal ions to produce the hydroxyl radical, which can oxidatively damage a range of proteins, lipids, DNA and RNA, and other biomolecules. What this tells us is that although the patient carries a "normal" MnSOD gene, the potential for oxidative damage is still increased. GPX1 has been implicated in the development and prevention of many common and complex diseases, including cancer and cardiovascular disease.9
Page 1, 2
|
![]()
![]()
![]()







