|
Page 1, 2
Introduction
Next-generation DNA sequencing: what does that mean and why would we use it?
Next-generation DNA sequencing (NGS) is a fairly new method for sequencing DNA segments in large numbers at a relatively low cost. NGS is considered by many as the second generation of DNA sequencing technologies and is sometimes referred to as massively parallel sequencing (MPS) due to the central technological advantage over the first-generation methods. It is useful for rapid and accurate characterization of nucleic acid sequences in living things. Application of NGS to infectious disease research and diagnostics will be significant for its use in rapid and accurate pathogen determination in viruses, bacteria, and protozoa.
Current clinical microbiological identification strategies generally rely on older technologies that have been in use for decades. One such technology is microbial culture, often used as a primary enrichment step that may take days. Culture, and by extension the majority of medical microbiology, assumes that a disease-causing bacterium is cultivable. Nonculturable organisms may be entirely missed as an etiologic agent, while emerging or unique organisms could easily be misidentified.1-4 Traditional alternative technologies include microscopy, serology, and, more recently, molecular-based assays. Molecular technologies commonly rely upon the amplification of pathogen specific DNA for detection. These tests are exceptionally sensitive; however, they can only detect a limited number of organisms or genetic variants. Recently, it is becoming increasingly clear that the majority of bacteria are fastidious (i.e., uncultivable) and present in polymicrobial communities. The true impact of uncultivable bacteria in medicine is likely significantly underestimated in addition to the ever-increasing need for more rapid diagnostic methods. These concepts have not gone unrecognized, and use of rapid culture-independent diagnostics has recently gained scientific and medical attention.
To address these challenges, a number of rapid microbial detection and identification systems have or are being developed. For cultured isolates, new protein based diagnostics such as matrix-assisted laser desorption/ionization time-of-flight (MALDI TOF) mass spectroscopy systems have been approved in Europe, with approval pending in the US. Bruker and BioMérieux have MALDI TOF systems that show promise. The IRIDICA system (previously named IBIS 5000) by Abbott is a rapid MALDI TOF mass spectroscopy PCR system and has been approved for use in Europe. Lastly, a number of point-of-care PCR based systems are available, with the BioFire system representing one of the most recent obtaining FDA approval. Generally, these systems require an initial culture step, rely on a limited reductionist approach, or have limited throughput.
Application
The diagnostic "holy grail" for both microbiologists and clinicians is an all-inclusive molecular approach that can be used in both acute and chronic infectious diseases. This perspective was reinforced at both the American Society of Microbiology and American Association of Clinical Chemistry conference meetings last summer. This view was again reiterated at the First ASM Conference on Rapid Next-Generation Sequencing and Bioinformatic Pipelines for Enhanced Molecular Epidemiologic Investigation of Pathogens last September. These emerging technologies and systems for infectious disease detection are rapidly becoming a reality.
Chronic diseases are also of great interest and represent an immense region of investigation. Examples of such diseases include chronic Lyme disease, chronic wounds, irritable bowel syndrome (IBS), sepsis, dental abscesses, chronic inflammatory disease, and fevers of unknown origin (FUO). Microbial involvement in these illnesses has been established or are suspected. Additionally, research suggests that these systems could have great relevance in the chronic inflammatory disease arena.
The application of molecular-based technology was well represented in recent Lyme disease research.5 This epidemiologic study of ticks in the San Francisco Bay Area utilized molecular-based approaches to assay for Borrelia species. This study demonstrated that 8.1% of the adult ticks harbor a Borrelia species and 1.8% have an emerging species, B. miyamotoi. This identification was made based on comparative sequence analysis and not by a matching control standard. In other words, the sequenced organism matched that of the textbook B. miyamotoi strain. Historically the "gold standard" for Borrelia detection has been antibody detection. Of course, these serologic techniques have limitations, such as antigenic cross-reactivity of a related, or sometimes distantly related, species. In sum, identification based on sequence identity in most cases is now considered a gold standard and is sufficient for detection and identification purposes.
Another rapidly expanding application of NGS-based methods is in stool microbiome analysis. NGS can allow for a comprehensive sequence based approach to studying and characterizing the plethora of bacteria in a patient's digestive system. Few systems currently exist that can accurately process samples and generate true enterotype results as most use culture or multiplex PCR, which can miss a number of important organisms. Furthermore, current methods suffer from severe limitations for the detection and identification of eukaryotic organisms including both protozoa and fungi.
Future iterations of diagnostics will rely heavily on molecular based technologies as they offer speed, inclusivity, and unparalleled accuracy. It is fortunate that these advantages are now being applied to infectious diseases enabling culture independent methods.3,4,6-34 These emerging diagnostic systems will share certain analysis methods that combine shared processing and bioinformatics processing pipelines.
System
The majority of NGS-based approaches will likely include similar steps:
1. Point of Care Sampling:Infected tissues, material, and/or fluid samples may be submitted for analysis. Proper collection techniques should be used to minimize contamination of the sample by nontargeted microbial populations. Blood draw sites should be cleaned thoroughly with disinfectants to remove potentially contaminating microbial DNA and cells. Tissue samples should be collected using aseptic techniques.
2. Nucleic Acid Extraction: Nucleic acid (DNA or RNA) content is then purified from the samples believed to contain the microbes. In the case of RNA it is converted into cDNA prior to sequencing.
3. Molecular Tagging and Amplification: The patient-derived DNA/cDNA is then prepared by fragmenting the full-length DNA into shorter segments compatible with sequencing. These fragments are often tagged with identifying sequences to allow for multiple samples to be pooled together. The subsequent DNA pool is then ready for NGS.
4. Next-Generation DNA Sequencing: Millions of digital DNA reads are produced by the NGS. The main NGS platforms include Illumina, Oxford-Nanopore, Thermo Fisher's Ion Torrent, and PacBio systems.
5. Bioinformatics Analysis: Software is used to sort and categorize the sequences depending on the intended down-stream use. Human DNA sequences are often removed from analysis to allow for focused processing of possible infectious agents. Analysis often utilizes heavy computational analysis to identify or recreate the genetic material of nonhuman organisms in the sample.
RIDI (Rapid Infectious Disease Identification)
Our laboratory has developed a metagenomic testing method that uses direct DNA sequencing and computational analysis to enable the detection, identification, and in the case of novel or divergent organisms, the identification of the nearest characterized microbial species. According to experts at the recent American Association of Clinical Chemistry (AACC) meetings, we are the first to enter the commercial marketplace of rapid NGS infectious disease diagnostics. This stands in stark contrast to the myriad of current indirect testing technologies, including serology, T-cell stimulation assays, and ELISA. Furthermore, this method allows us to provide a relative measure of the bacterial, protozoal, or fungal contribution and diversity within a given sample. In summary, this method aims to identify the genetic composition and diversity across all microbes in a sample, simultaneously.
Our laboratory uses the Ion Torrent PGM system, clinically regulated methods, proprietary reagents, and in-house bioinformatics analysis to identify and survey the organisms of an unknown or polymicrobial infection. We call this system the Rapid Infectious Disease Identification system, or RIDI. It is important to note that the RIDI bioinformatics system has proven compatibility across other NGS systems, including the Illumina and PacBio platforms. By using direct DNA sequencing from the Ion Torrent PGM coupled with computational analysis, we can identify or match a detected organism to the nearest microbial relative in a given clinical sample. Furthermore, this method allows us to provide a relative measure of the microbial contribution and diversity within a given sample. Fry Laboratories believes that wider adoption of this test in clinical use will have far-reaching implications by not only providing superior, unbiased, sequence-based diagnosis, but also reducing patient mortality, morbidity, length of stay, and associated hospital and health care costs. This test is currently offered worldwide and is performed as a Laboratory Developed Test (LDT) in our CLIA regulated diagnostics laboratory.
Validation of Next-Gen Systems, or, How Well Do These Systems Work?
NGS manufactures and industry experts agree that there is need for comprehensive validation and standardized control procedures in the NGS diagnostics field. Our Pan-Bacterial Metagenomics assay by RIDI has successfully and repeatedly sequenced Borrelia burgdorferi and several Bartonella species standards from the American Type Culture Collection (ATCC). Additionally, this assay has an ever-increasing list of validated and control ATCC species and taxonomic groups that include Acholeplasma laidlawii, Acinetobacter baumannii, Bacillus cereus, Bartonella bacilliformis, Bartonella henselae, Bordetella pertussis, Borrelia burgdorferi, Capnocytophaga gingivalis, Clostridium difficile, Enterococcus faecalis, Escherichia coli, Gemella haemolysans, Haemophilus influenzae, Klebsiella pneumoniae, Legionella pneumophila, Mycoplasma arthritidis, Mycoplasma fermentans, Mycoplasma genitalium, Mycoplasma hominis, Mycoplasma pneumoniae, Ralstonia solanacearum, and others.
Our laboratory has undertaken the mapping of bacterial, protozoa, fungi, and simple eukaryotic populations in a variety of sample types, including stool, blood, joint fluid, abscess, cerebrospinal fluid, bone, and tissues. This is especially true in certain disease states such as IBS, chronic fatigue syndrome (CFS), Lou Gehrig's disease, multiple sclerosis, atherosclerosis, and prostate cancer.
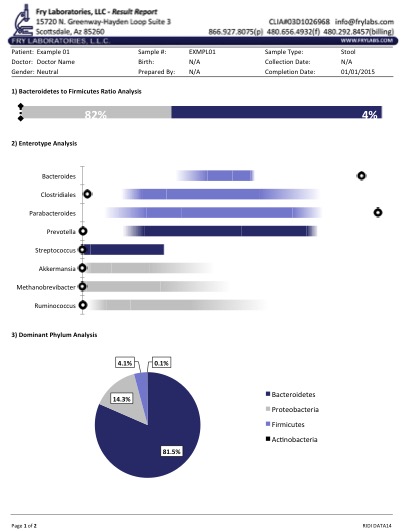
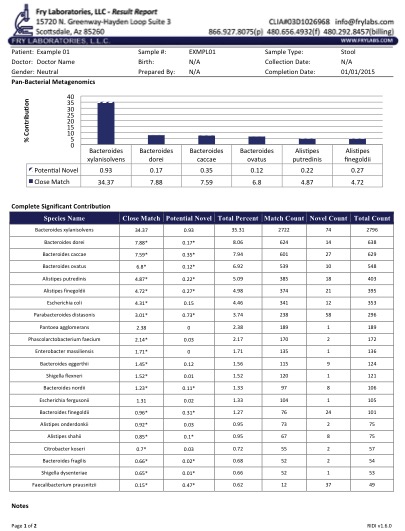
Figure 1 displays a typical report in a patient with IBS. Of note is the identification of pathogenic Shigella spp. when simple multiplex PCR and culture techniques had failed. Our ongoing research with next-gen detection systems has allowed us to verify a variety of aquatic "parasites" in humans.
Page 1, 2
|
![]()







